
Bonus Breed – Samoyeds!
The Samoyed population is a “bonus” breed! Their profile was made not through community effort, but as a result of a study published by Dr. Niels Pedersen last year, which not only analyzed the breed’s biodiversity, but also identified a recessive mutation for enamel hypoplasia.
The Samoyed is a breed developed for use by the Nenets (Samoyede) peoples of Northwest Russia and Siberia. The modern day breed was established by a few individuals who imported dogs to develop the breed in the UK. This establishes a very small initial genepool for this breed, but what did UC Davis Veterinary Genetics Laboratory and Dr. Niels Pedersen discover in the genetic analysis of the breed? Link to full report here.
Inbreeding – how closely related are the parents of the typical Samoyed?
Samoyed breeders have mostly as a whole randomly bred their population. However, a proportion of the population is quite inbred, which is offset by the part of the population that is outbred. From the report:
One half of the dogs had IR values over 0.042 and one quarter had values of 0.132-0.503. A value of 0.25 would be average for a litter of puppies born to parents that were full siblings in a random breeding population. An IR value of 0.498 would only occur if the full sibling parents were more highly related to each other that the average dog in the population. This highly inbred quarter of the population is balanced by the one quarter of outbred dogs with IR values of -0.276 to -0.056 to This balancing of highly outbred and inbred dogs is why the Ho, He and F values calculated for the whole population using allele frequencies of the 33 genomic STR loci gave the impression that the population of 190 dogs tested were being randomly bred.
It is fantastic that a part of the population is so outbred – meaning that the genetics from the sire and dam are not the same – however a good portion of the population is highly inbred. An inbreeding measurement, or IR, of .15 or higher is considered very inbred. This population could benefit from using breeding software to ensure that their breeding mates are actually unrelated as they might think (or that their linebreedings are not too close).
Breed wide diversity
Biodiversity and allelic richness are words we try to discuss regularly at BetterBred. Namely, how many different versions of genes or alleles are there? The breeds with more allelic richness, that are also well distributed, tend to be healthier. What did they find about the Samoyed population?
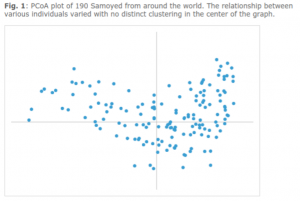
The plot shows that the 190 dogs cluster as a single breed, although the plot is somewhat diffuse with several more distant outliers. This suggests that there is still phenotypic, and therefore genotypic, variation in the breed.
What does this mean? This population does not appear to have a genetic bottleneck around which most dogs would cluster, and there is a presence of genetic outliers. Breeders should seek out the presence of the outliers in order to increase another measure we discuss: two terms called average alleles per locus and effective alleles per locus. The closer the effective alleles (we call these “alleles that are effectively contributing to your population”) are to the average alleles, the better distributed the breed. Why would you want a well distributed breed? It eliminates risk of loss of diversity due to genetic drift and it dilute disease genes that might be concentrated if there were a bottleneck.
From the report:
The population of Samoyed, when examined as a whole population, had a mean number of alleles (Na) of 6.455 across all 33 genomic STR loci. The mean effective alleles (Ne) per locus were 3.249. This means that about one half of available alleles accounted for most of the diversity. The Na and Ne values for Samoyed are lower than genetically diverse breeds such as Miniature and Toy Poodles, similar to Labrador and Golden Retrievers, somewhat more than Flat Coated Retrievers and much greater than for Swedish Vallhund.
Each breed should make an effort to raise their effective alleles per locus. How do you do this? You use BetterBred’s breed management software to breed for higher than breed average Outlier Index (OI). This measurement was created to help breeds redistribute diversity and maintain what they have as a breed.
Diversity compared to the Village Dogs
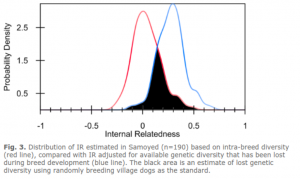
Dr. Pedersen’s hallmark comparison is to a population of village dogs that has a great amount of genetic diversity. They compare the results of each breed to the results of the village dogs to estimate the diversity remaining in each breeding population.
The dark area of overlap between the IR and IRVD curves is an estimate of the amount of genetic diversity in modern village dogs that is still present in Samoyed (i.e., 35%). This is lower than the 60% or so retained diversity seen in genetically diverse breeds such as the toy poodle or 54% in Labrador Retriever, but higher than the 7% observed in Swedish Vallhund, 15% in Doberman Pinschers, or 27% in Shiloh Shepherd.
This breed has an average amount of diversity of the breeds that thus far have been tested. Great care should be taken to preserve what exists in the breed currently.
DLA – a Look at the Immune system
The DLA is the region of the dog’s genome that controls the immune system. The canine diversity test at UC Davis identifies the most regions of the DLA of any commercial test out there currently. It identifies both Class 1 and Class 2 DLA haplotypes. They label them as numbers, but these numbers do not change from generation to generation. A puppy receives one class 1 and one class 2 DLA haplotype from each parent. 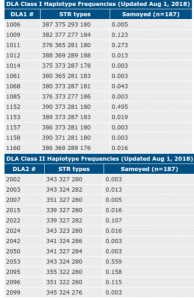 You can read more one DLA and its use in a breeding program here.
You can read more one DLA and its use in a breeding program here.
Considering that this breed was founded on imports by a few people in the UK, there is a great variety of DLA haplotypes. However, from Dr. Pedersen’s report, “The 1152 and 2053 haplotypes were found in 50-56% of individuals, respectively“. Other DLA haplotypes that are have higher representation in the population include 1009 and 1011 (they represented 12.3% and 27.3% of the tested class 1 DLA haplotypes) and 2022, 2095 and 2096 (10.7, 15.8 and 11.5% of the tested class 2 DLA haplotypes).
From the report:
The 2053 haplotype was found at high frequency in Swedish Vallhund (0.49), Flat Coat Retriever (0.126), and Shiloh Shepherd (0.317), but not in association with 1152 (Table 7). Several other class I and II haplotypes found in Samoyed were shared at low frequency with other breeds, Labrador Retriever, Golden Retriever, Havanese, Miniature Poodle, Standard Poodle, and Swedish Vallhund. Unfortunately, no comparative data was available for Nordic breeds such as the Siberian Husky and Malamute.
This means that there is one DLA haplotype that is most represented in the population. There are a few others with good representation and there are others that are much less well represented. The one DLA haplotype that is found in the majority may be due to a single founder who contributed to the breed’s formation or a bottleneck at some point within the breed at a time like WWII. Breeders should make an attempt to breed for heterozygous DLA and seek out the less common DLA types to maintain what they have thus far, to avoid losing their DLA diversity through genetic drift.
Conclusion
From Dr. Pedersen’s report:
The Samoyed breed has average genetic diversity among the breeds that we have studied based mainly on the Na and Ne values, IR/IRVD curve overlap and DLA class I and II haplotypes. The breed has less genetic diversity than Miniature, Toy and Standard Poodles, Italian Greyhounds, Golden Retriever, Labrador Retriever, Havanese, Akita and Alaskan Klee Kai; similar genetic diversity to Black Russian Terriers, Flat Coated Retriever, Doberman Pinscher and Giant Schnauzer; and greater genetic diversity than Shiloh Shepherds and Swedish Vallhunds.
This breed has sustained an average amount of genetic diversity compared to other breeds, and the genetic diversity appears to be fairly well distributed for what they have. However, the DLA haplotypes are disproportionately distributed within the breed.
Our Recommendation
Breeders should strive to maintain the genetic diversity that is still present within the breed, as well as seek out less related mates to keep down inbreeding. When all else is equal, seek for the less represented DLA haplotypes and heterozygous (two different kinds) of DLA haplotypes. There is a reported presence of outliers according to Dr. Pedersen (see the PCoA graph above), so those should be sought in order to distribute the genetic diversity of the breed more equally. Breeders should strive to breed for higher than average OI, lower than breed average IR (inbreeding) in order to raise your breed’s effective alleles per locus.
You can order your test here today and hit the ground running with our revolutionary breed management software!

 Previous Post
Previous Post Next Post
Next Post


