
Let’s talk about COI and other estimates of inbreeding
COI, “coefficient of inbreeding,” is a very specific concept. We breeders have to pay attention to the definitions given by different people and companies.
Pedigree based COI is a specific formula developed in 1922 by Sewell Wright, one of several prominent mathematically minded scientists who together established the science of population genetics. Even back then they had some disagreements on theory, but Wright’s formula ended up as the most popular and it’s the one most of us use today. Specifically, it is a pedigree-based statistical calculation designed to give us a probability that an animal will inherit the exact same gene, called homozygous, from the exact same ancestor.
So let’s imagine a scenario. DNA mutates occasionally when sperm or eggs are created, sometimes with dramatic effects. So let’s imagine a dog who had a unique, never-seen-before glitch in his DNA that created a mutation on a single gene for the dreaded Exploding Head disease. Let’s say that’s an autosomal recessive disease, so he has one normal gene and that one hidden recessive mutation. Of course, heads only explode when a dog inherits two copies of this precise, unique mutation, so this dog is perfectly healthy. He’s the only dog on earth with this exact mutation, and if he’s never inbred on, no one ever inherits two copies and no heads explode.
But let’s say he’s a magnificent MBIS dog with all kinds of wonderful qualities. All his kids are fabulous. Everyone is happy. 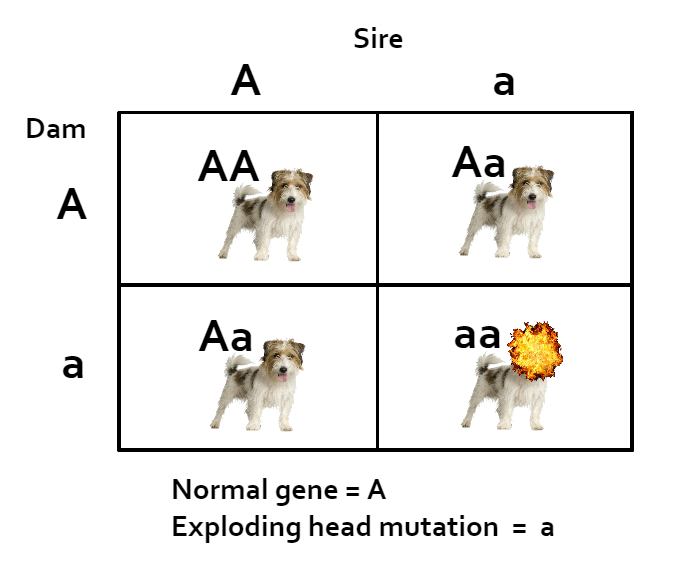
Then someone doubles up on him – she breeds half siblings together who both have him as a sire. Some of his double grandkids are as fabulous as he is. Some of them inherit TWO of the exact mutation from both their sire and their dam and… their heads explode.
That’s an extreme example (obviously!) but let’s say no one doubles up on him until he’s in the 4th generation on both sides a couple of times. There’s a calculable mathematical probability that his descendants will inherit two copies of that mutation (and their heads will therefor explode.) That mathematical probability is the coefficient of inbreeding, COI.
Often in purebred dogs there are no repeats of ancestors on both sides of a pedigree in the first 5 generations and breeders think dogs are unrelated. Sometimes there are no repeats until the 10th generation. But in most breeds,at the 15th or 20th or 30th generation, there are the same individual dogs over and over and over, and on both sides of every pedigree.
That’s why a 5 generation COI can be very low and a 10 generation COI can be much higher and a 20 generation COI can be sky high. All breeds have ancestors in common – it’s how breeds were created. How many they have in common, and how many generations are used to calculate a pedigree based COI matters. What’s more, pedigree based COIs also assume that the very first generation it considers, whether the 5th generation or the 45th, are the “founders,” and are completely unrelated. This is rarely the case. This is the inherent flaw in any pedigree based management system. It is only as good at the pedigree is deep, complete and accurate.
Breeders often think that in 20 generations DNA will have changed a lot – mutated, recombined, something. Twenty generations is a very long time for dogs and humans, but it’s actually a blip in evolutionary time. Genes don’t really change that much in that many generations, especially when humans are selecting for all the same traits for 20 generations, which may skew the gene pool toward more and more similar animals. Add to this that stud books are closed – and the only variations that can be added are mutations, which happen slowly in a human timeframe. For the most part closed breeds have a finite set of genes and their versions – versions which take often thousands of years or generations to develop. So while we breeders think we are developing better ear set or tail set or coat texture or angles front and rear, we are really just selecting which of the gene version we prefer. And every time we breed one gene out on purpose, so it no longer exists in the breed, we are also eliminating a lot of other genes inadvertently as well.
One way we can assess how much we’ve done this already – or not – is to assess the inbreeding levels in a dog, a line, or a breed.
Different DNA methods are better at assessing inbreeding in different ways. “Genetic COI” is the term one popular company has coined to describe their estimate of inbreeding, but it’s really just a common method of using what are called “runs of homozygosity” in DNA. It is not really comparable to pedigree based COI because it estimates inbreeding by sampling actual homozygosity in a fairly small fraction of the actual canine genome. Then it extrapolates that homozygosity to say the whole genome is likely to have the same percentage of homozygosity as the sampled DNA. This is also an estimate – a decent one. However, the canine genome is 2.8 billion base pairs and the chip they use records about 230,000 of those base pairs. Each point of data is called a locus – plural is loci – and again, that simply means a location on the DNA. Loci can be big or small – it’s a general term, but with the technology that company and others use, SNPs, loci are very tiny and each locus is either homozygous or heterozygous. That 230k sampled loci are on average 12,000 base pairs apart. If they find that many of these sampled loci in a row are all homozygous, this is called a “run of homozygosity,” or ROH, and they assume the whole section is homozygous. The more inbred a dog is, the more of these ROH they will have.
One reason researchers like this method is that it assesses all inbreeding, including ancient inbreeding, and not just recent inbreeding. So that’s one reason that company’s “Genetic COI” can be much higher than a typical pedigree based COI, which is limited to 5 generations, or 10 or 20, depending on the depth of the pedigree.
The real question is, what does this mean for breeders? Do we need to know more recent inbreeding or all the ancient inbreeding as well?
Since most recessive mutations arise in the manner described in the Exploding Head disease example above, they are almost always very highly breed specific. Pedigree based COIs can be reasonable for assessing risk for recessive disease within breeds, as long as they are calculated back to the founders of the breed, and since this is rarely done or even possible, DNA based estimates of inbreeding have been shown to be far more accurate.
Ancient inbreeding however, is not so useful. Most obvious recessives tend to be at very low frequencies (meaning there are very few if any in each breed) due to “purging” or selective breeding over the many generations by human selection, so the diseases that are found in more than one breed tend to be complex, and COI is not useful for assessing risks for complex disease.
Occasionally two similar breeds may have the same recessive diseases, in which case ancient inbreeding estimates may be useful, but this is rarely the case. They might both have, say, the MDR1 gene in common if they are both herding breeds, but that’s a single gene in a huge genome, for which there is a DNA test. Better just to test for that disease.
A pedigree based COI for a cross between two very similar breeds would of course be 0%, and since most recessive diseases would be different in each breed, this 0% COI translates to a useful recessive disease prediction. For the most part, however, such a cross would create highly heterozygous puppies – and horrify most breeders. The “Genetic COI” of puppies from a cross of two similar similar breeds would likely not be zero because they’ll have a lot of very old ancestry in common, but it will also not be very meaningful from a breeder’s perspective.
Another popular company uses a straight homozygosity estimate and adds that to weighted risks for known diseases to come up with an index that is meant to predict genetic health. They also use SNPs, and an estimate of homozygosity. However – no matter which company does it with any method – estimates of inbreeding are fairly similar when compared breed to breed. They all are able to identify highly inbred dogs and overall inbreeding in breeds. Breeders, however, don’t actually need precision in knowing how inbred a dog is – very or somewhat or not very inbred may be enough – as much as they need to know what different levels of inbreeding may mean for breed specific disease risk in their breeding program and the puppies they produce.
The UC Davis canine diversity test that BetterBred bases their analyses on uses highly specific sections of DNA called microsatellites which are very precise and good for assessing more recent inbreeding, like from 20 to 100 generations – but not the ancient ones. Their method is very precise for individuals, and are the same kind of DNA tests as those used in human forensics to catch criminals and to establish paternity and more distant relationships. Much of the canine panel of microsatellites used by UC Davis was selected based on human standards for precision because they are used in US and international courts – the rest are loci identified by ISAG, the International Society for Animal Genetics, for use in parentage testing. Microsatellites assess homozygosity in a very different way than do the SNP chips used by other companies. Instead of taking a sample of an individual dog and extrapolating homozygosity, they estimate how similar the genetics are that a dog inherited from each parent, using a widely used population genetics calculation called “internal relatedness” or IR, as well as other calculations. This entire method is completely different from how SNPs work.
Most breeders are only familiar with the idea of homozygosity or heterozygosity, because that’s what’s usually emphasized, and that’s what COI implies. Diversity considerations, however, really should include more than that. With a locus on a SNP chip there are only two possible versions of markers, let’s call them A and B – and therefore, since each dog inherits one version from each parent, there are only three possible outcomes: AA, BB, and AB. That’s it.
With a microsatellite, which is a much larger chunk of DNA, there can be anywhere from, say, 3 to 24 versions. The more versions there are at each locus, the more powerful the test is. These loci are very carefully chosen specifically for their many versions.
So let’s imagine a locus in a breed, and let’s say there are ten different versions found in at that particular locus. This in itself important information, because breeds that have suffered a loss of diversity might have 5 versions at that locus, whereas genetically diverse, healthier breeds might have 15. Let’s call the 10 versions at our imaginary locus A, B, C, D, E, F, G, H, I, and J. Each dog inherits one version from each parent and so possible combinations of those at that locus would be AA, AB, AC, AD, AE, AF, etc., all the way to JI, JJ. So 100 different combinations, rather than 3. Each combination has a particular mathematical likelihood of appearing in a dog in each breed and so this is how it offers very precise probabilities on relatedness between dogs.
Then there is the frequency of the versions – how often each version appears in a breed. With 10 versions, the probability of a locus being homozygous can be quite small, but usually some versions are much more common than others. So let’s say version A is very common in the breed but version J is very uncommon. If a dog is homozygous for A, that doesn’t say much about how homozygous genome wide that dog is because lots of dogs carry A. But a dog that’s homozygous for J inherited the same rare version from both parents, which means the parents must have ancestors in common. When parents are related, they have inbred puppies. That JJ, therefore, is a more meaningful homozygous pair than an AA at the same locus. It also has a mathematically calculable probability. So that is also taken into consideration.
And that’s just the information at a single microsatellite locus. Multiply that information times the 33 microsatellites on the UC Davis Canine Diversity test, and you have an exponentially large amount of information about the dog in question – as much or more, although different information, than what you get from a SNP chip like other companies use. Multiply that dog’s information by 100 dogs in the same breed and you will know an enormous amount about the breed. You just have to be able to analyze that data properly and know how to interpret it.
This incredible precision and probability is why you only need 13 carefully selected microsatellite loci to identify any human on earth with 99.99999% probability. Humans aren’t usually inbred, so fewer well selected markers work. Dog breeds are inbred so they need more, but UC Davis found that 33 markers gives them ample accuracy in all breeds.
So this is why we can’t compare the different kinds of markers one to one, other companies’ methods and UC Davis. Breeders don’t know that. Geneticists all do.
We also can’t compare the results of a single dog from one test to the next. And we can’t compare an individual’s result in one of those to its pedigree based COI.
We can, however, compare the results of an individual tested by a single company to the breed-wide results from that company. But again, we need to know what that tells us. Does high homozygosity in a breed indicate disease? Sometimes. Not always. Does low homozygosity in an individual dog indicate low risk for disease? Sometimes. Not always. It depends on the disease and the breed.
So try to remember that everyone is dipping their toes into some complex concepts here and there are lots of people giving opinions without a full understanding of what it means for breeders.
My advice is to ask that question a lot. “Ok great. What does that mean for me as a breeder?”
“What does this inbreeding estimate indicate about the health, breeding value and prospects of my dog individually and in relation to his breed?”
And make sure you get concrete answers.
 Previous Post
Previous Post Next Post
Next Post


